CITA ESTE TRABAJO
Diego Martínez R, Cano de la Cruz JD, Sánchez Sánchez MI, Vázquez Pedreño LA, Jiménez Pérez M. Development and training of a convolutional neural network for the detection of esophagitis in endoscopic images. RAPD 2025;48(2):xx-xx. DOI: 10.37352/2025483.1
Introduction
Digestive endoscopy can provide a direct and minimally invasive assessment of the gastrointestinal tract; however, the assessment remains subject to some inter-operator variability, resulting in significant differences in the efficiency of the technique depending on who performs it.
In this context, artificial intelligence (AI) and, in particular, convolutional neural networks (CNNs), have emerged as promising tools to improve the accuracy and reproducibility of endoscopic diagnosis. These deep learning architectures, inspired by the way neurons in the human brain interconnect, are composed of multiple layers of artificial neurons that are trained to recognize patterns from data. During training, each layer extracts features or characteristics of increasing level of complexity, constantly adjusting the weights of their connections in order to improve their classification or detection ability. In current clinical practice, AI has begun to be used in the detection of colorectal polyps, characterization of gastric lesions and other applications that seek to support endoscopic diagnosis.[1],[2].
The present work focuses on the development of a convolutional neural network specifically trained for the detection of esophagitis in endoscopic images. Through the use of image processing and deep learning techniques, we seek to improve the automated diagnostic capability. This approach not only has the potential to improve clinical decision making, but also to streamline workflow in medical settings, facilitating faster and more accurate diagnosis for patients.
In this article, the process of developing and training the AI model, as well as its validation using an endoscopic imaging dataset, will be described. In addition, the challenges and future perspectives in the integration of these systems into clinical practice will be discussed, with the aim of improving the quality of endoscopic diagnosis and care for patients with esophageal diseases.
Subject Matter and Methods
In this project, the Python programming language together with the Keras and TensorFlow[3] software libraries were used to implement and train a deep neural network architecture. The working environment selected was Google Colab Pro, which provides access to powerful graphics processing units (GPU), in this case an Nvidia A100. This is essential to significantly reduce training times and to be able to handle large volumes of image data.
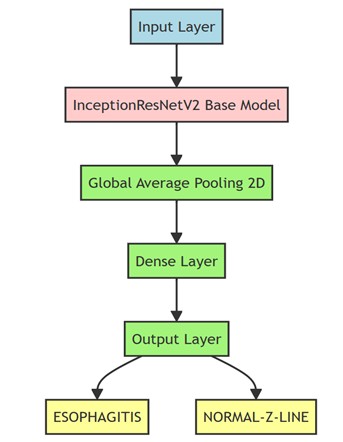
The base architecture chosen was InceptionResNetV2[4], originally developed by Google researchers and trained on the massive, public ImageNet[5] dataset. IInceptionResNetV2 combines the advantages of the convolutions of the Inception family with the stability and efficiency of residual-type connections, resulting in a model that maintains an appropriate balance between accuracy and training speed. The original InceptionResNetV2 model, after being trained on the classification of thousands of ImageNet categories, was adapted for our specific use case. For this purpose, the output layers were replaced by custom layers designed to perform binary classification. In particular, three dense (fully connected) layers were added, culminating in an output layer with the ideal activation (to distinguish between two classes: esophagitis versus a normal Z-line (see Figure 1).
Figure 1
Scheme of the model created, in green the layers added to the InceptionResNetV2 base model shown in red.
The fine-tuning process was carried out by keeping the initial layers of the model - those responsible for extracting features - fixed and retraining specific final layers. In this way, we took advantage of the richness of the weights obtained from ImageNet and oriented the network towards the discrimination of our two clinical categories of interest. This allows a much more efficient use of data and training time by avoiding training the model from scratch.
We trained the model with the KVASIR endoscopic image set[6], a repository covering digestive endoscopy images. In particular, we used 2000 images corresponding to both normal Z-line and different degrees of esophagitis, dividing the data into 80% for training and 20% for validation. This balance in data partitioning allowed robust training of the model and a reliable preliminary assessment of its performance. To perform a more thorough validation, we employed the HyperKVASIR image set
[7], totaling 1164; 932 normal Z-line and 232 esophagitis. At this stage, milder cases were excluded, focusing only on grades B, C and D of the Los Angeles classification, in order to assess the ability of the model to identify more advanced lesions. Additionally, a third set of images from the Regional University Hospital of Malaga was incorporated. This set consisted of 203 esophagitis images (all of them pathological) that included different degrees of severity (76 images of Los Angeles grade A, 42 of grade B, 28 of grade C, 22 of grade D and 18 in the category of others intended for when the endoscopist did not specify the degree of esophagitis) thus reinforcing the diversity and clinical representativeness of the data used in the study (see Table 1).
Table 1
Table summarizing the data sets used in the project.
| Image set | Number of images | Description | Use |
| KVASIR[6] | 2000 | Images of normal Z-line and different degrees of esophagitis. | Training and validation of the model. |
| HyperKVASIR[7] | 1164 | Images of normal Z-line (932) and esophagitis (232) | Evaluation of model performance in advanced lesions. |
| Regional University Hospital of Malaga | 203 | Images of esophagitis with different degrees of severity. | External validation in a real clinical setting. |
Results
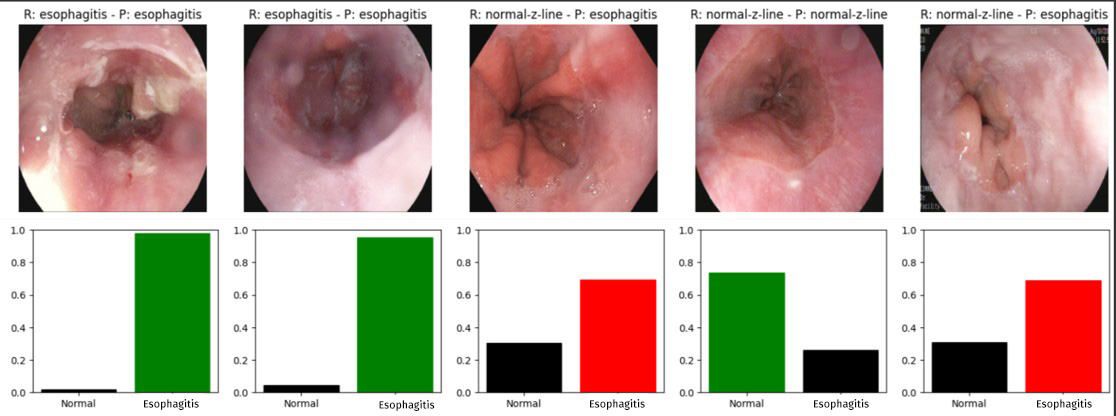
A detailed example of the individual predictive ability of our model is presented below, showing the confidence percentages assigned to each diagnostic category. To illustrate this aspect, we have randomly selected five images (see Figure 2 ). When evaluating the detection in the image sets, the following was observed. In the KVASIR set-corresponding to the 20% of images reserved for evaluation and not used during training-of the 200 images that corresponded to normal Z-line, the model correctly identified 164, while 36 were misclassified as esophagitis. Similarly, of the 200 images that actually corresponded to esophagitis, 153 were correctly classified, and 47 were mistaken for normal Z-line (see Table 2 ). On the other hand, in the HyperKVASIR image set, the confusion matrix revealed that, of 932 normal Z-line images, the model correctly classified 833 and failed in 99 cases. On the other hand, of the 232 images corresponding to esophagitis, 216 were correctly identified, while 16 were misclassified as normal Z-line (see Table 3 )
Figure 2
Representation of the individual predictions made by our model. Above each image is shown the actual label (R:) and the predicted label (P:) and below a bar chart with the confidence percentage assigned to each class in the prediction which is colored green if it is correct and red if it fails.
Table 2
Confusion matrix of the prediction of the 400 images of the KVASIR image set corresponding to the 20% of images we have reserved for evaluation.
| KVASIR | Z line Normal (Predicted) | Esophagitis (Predicted) |
| Z line Normal (Real) | 164 | 36 |
| Esophagitis (Real) | 47 | 153 |
Table 3
Confusion matrix of the prediction of the 400 images of the HyperKVASIR image set.
| HyperKVASIR | Z line Normal (Predicted) | Esophagitis (Predicted) |
| Z line Normal (Real) | 833 | 99 |
| Esophagitis (Real) | 16 | 216 |
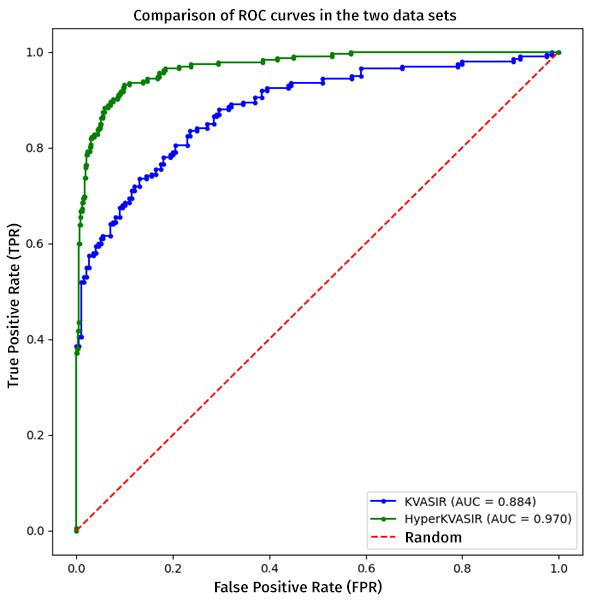
Figure 3
Representation of the ROC curves in the validation with KVASIR (which includes esophagitis of all degrees of severity) and HyperKVASIR (which only includes Grades B, C and D, discarding the mildest cases) in which a better curve is observed in the latter set.
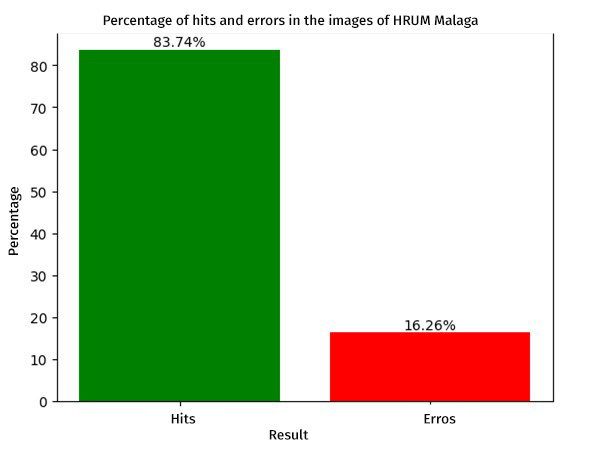
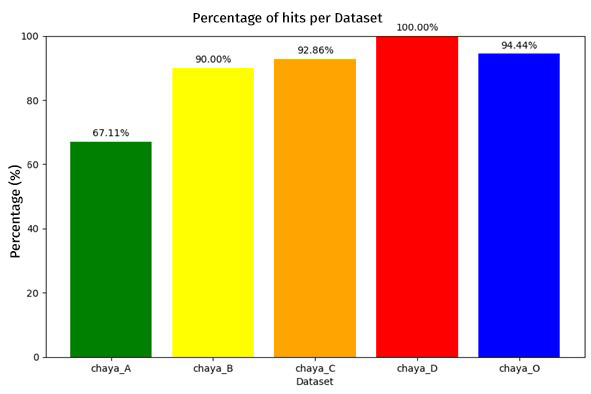
To compare both evaluations we will use the ROC curve metric, which is essential to determine the overall performance of our classification system. A relevant aspect was the exclusion of Grade A esophagitis only in the HyperKVASIR database, in order to evidence that by having a higher contrast between the control image and the pathological image, the model can discriminate more effectively, which favored the efficiency in pathology detection, achieving area under the curve (AUC) values of 0.884 for the KVASIR dataset and 0.970 for the HyperKVASIR dataset. These results point to a high level of diagnostic accuracy (see Figure 3). Likewise, the graph below illustrates the percentage of hits obtained by the model when evaluated with the cohort from the Regional Hospital of Malaga. This independent analysis is essential to corroborate the applicability of the model in diverse clinical settings and to validate the robustness of the methodology in real circumstances (see Figure 4).
Finally, when classifying performance according to esophagitis severity, a positive correlation was observed between the increase in severity and the percentage of hits. This finding suggests that the algorithm is particularly efficient in detecting more advanced lesions (see Figure 5).Discussion
The findings of this study highlight the potential of the InceptionResNetV2 architecture for the detection and classification of esophagitis based on endoscopic images. The use of pre-trained layers with the extensive ImageNet dataset, along with fine tuning focused on the esophagitis vs. normal Z-line problem.
AUC (area under the ROC curve) value obtained on the two main validation sets - KVASIR and HyperKVASIR - provided evidence of both the consistency and generalizability of the model. The exclusion of milder grades of esophagitis (Grade A) in HyperKVASIR showed how greater contrast between normal and pathological images facilitates sharper discrimination, reinforcing the hypothesis that the model performs particularly robustly in more severe lesions. This aspect becomes clinically relevant, given that, in practice, advanced lesions often require more timely diagnosis and treatment.
Independent analysis on the image set of the Regional University Hospital of Malaga provides further evidence of the applicability of the approach in a variety of settings. The results confirm the usefulness of the methodology not only in public databases, but also in a real clinical setting, with variations in imaging conditions, types of endoscopic equipment and population characteristics. The fact that the performance of the model increases in relation to the degree of severity of esophagitis suggests that the neural network is able to detect more accurately the most evident structural alterations. However, further classification of incipient lesions is necessary, as early identification is essential in medical practice to prevent future complications and improve the prognosis of the condition.
Despite the promising results, this study has some limitations. On the one hand, the total number of images, although significant, could be expanded to cover a greater representativeness of the different forms of esophagitis presentation, especially those of grade A. On the other hand, factors such as variability in image quality and the presence of artifacts during endoscopy may influence the accuracy of the model.
Conclusions
Although the usefulness of this model is not yet applicable to clinical practice, this study highlights the potential of deep neural networks in endoscopy and underlines the importance of collaboration between hospitals to create multicenter databases. Increasing both the number and diversity of images is crucial to train models that, in the future, can be implemented in more relevant clinical contexts. The results obtained here highlight that, although the model shows high sensitivity in advanced lesions, challenges are still faced in the identification of incipient stages and in the adaptation to different imaging conditions. Practically speaking, multicenter validity, the use of data augmentation techniques and the integration of these systems into clinical workflows could, in the long term, favor more agile and accurate diagnoses for a variety of gastrointestinal pathologies.

 Descargar número completo
Descargar número completo Download full issue
Download full issue