CITA ESTE TRABAJO
Muñoz-García-Borruel M, Álvarez-Barrientos A, Muñoz-Sanz A, Gutiérrez-Martín Y. Tetraspanins and miRNAs in urinary extracellular vesicles in patients with colonic polyps and a family history of colorectal cancer. RAPD 2025;48(4):127-133. DOI: 10.37352/2025484.1
List of abbreviations
FH CRC: family history of colorectal cancer.
CRC: colorectal cancer.
FC: flow cytometry.
SD: standard deviation.
miRNA: microRNA.
qPCR: quantitative real-time PCR.
IQR: interquartile range.
EVs: extracellular vesicles.
Introduction
Colorectal cancer (CRC) ranks third in incidence and mortality in both sexe[1]. Thanks to early diagnosis and the implementation of population screening programs, mortality has decreased. However, due to lifestyle habits and lack of resources in health systems, CRC remains a major public health problem. Although most CRC are sporadic, about 15% have a family history of CRC (FH CRC)[2]. Therefore, early diagnosis of preneoplastic lesions is mandatory in individuals with first-degree FH CRC.
A current trend is the search for non-invasive tests aimed at diagnosing patients with precursor lesions or stratifying risk groups. These include biomarkers (liquid biopsy) that can be analyzed in a multitude of biological fluids. Urine is an excellent sample because of its ease of collection and storage and its abundance of metabolites and other molecules. One way to approach this type of research is the study of extracellular vesicles (EVs) and the molecules they carry. EVs are very abundant in the organism and constitute intercellular information vehicles[3],[4].
Within the cargo of EVs, microRNAs or miRNAs and some membrane proteins, tetraspanins, stand out as markers of EVs, since in addition to being part of the membrane structure, they participate in the genesis, distribution, transport and elimination of the components of the cargo[5]-[8].
A multitude of miRNAs have been identified that appear to be involved in the development of polyps and CRC[9]. The miRNAs may have oncogenic (onco-miRNAs) or anti-oncogenic (anti-oncomiRNAs) activity[10]-[12]. Onco-miRNAs include: miR-31 (KRAS stimulator), miR-21, with oncogenic properties through repression of the PDCD4 (Programmed Cell Death 4) target (proinflammatory tumor suppressor), and miR-200, important in the maintenance of epithelial identity that represses promesenchymal transcription factors and promotes metastasis. On the other hand, among the miRNAs with anti-oncogenic functions, the following stand out: the miR-34 family, key in the repression of tumor migration, invasion and metastasis formation, and the miR let-7 family, included among the most important in the repression of oncogenesis due to its abundance and antiproliferative function.
Compared to liquid biopsy, urine is an excellent biological fluid that offers important advantages for its analysis: it is an affordable and convenient sample for the researcher and the patient, reproducible, little studied and abundant in EVs and miRNAs from different origins of the organism, including the colon.
The objectives of this work were to analyze the tetraspanin profile by flow cytometry (FC), to establish urinary miRNAs that could be useful as biomarkers of neoplastic or preneoplastic colonic pathology, and to demonstrate the practical value of urine in the analysis of EVs and intravesicular cargo.
Patients and methods
This was a prospective analytical observational study involving subjects undergoing colonoscopy for AF first-degree CRC aged 18-70 years and subjects who underwent screening colonoscopy (control group). The study was conducted between November 2019 and November 2020. All patients had a urine sample collected on the day of colonoscopy.
Exclusion criteria were: age <18 years or >70 years and persons with endoscopic findings of infectious/ischemic colitis, inflammatory bowel disease, or CRC.
The colonoscopies had to meet the recommended quality criteria. The resected polyps were sent to the Anatomic Pathology service of each center..
On the day of the colonoscopy, a urine sample (10-20 ml) was extracted and stored in a freezer at -80ºC until analysis. Subsequently, the samples were sent to the laboratory of the Applied Bioscience Techniques Service of the University of Extremadura for analysis by CF and massive sequencing. The selected samples were identified by a numerical code.
The data of each patient were archived with appropriate security measures in compliance with the Organic Law 15/1999 on Data Protection.
Statistical analysis
Qualitative variables were presented using frequency tables and quantitative variables were summarized as mean and standard deviation (SD) or median and interquartile range (IQR) for variables that did not follow a normal distribution. IBM SPSS© v. 20.0.0 was used.
Laboratory methodology and bioinformatic analysis
Urinary samples were centrifuged and filtered to obtain EVs making two aliquots: one for the study of tetraspanins by CF analysis and the other for the extraction of miRNAs and their massive sequencing using Ion Torrent technology.
For the study of tetraspanins in EV membranes, staining with Carboxyfluorescein Succinimidyl Ester (CSFE) was performed to analyze the quality of the preparation and the other sample was used for staining with the antiCD9, antiCD63 and antiCD81 antibodies conjugated with the different fluorochromes and thus visualized by the corresponding channels of the flow cytometer (Cytoflex S, BeckmanCoulter).
For the sequencing of miRNAs, we first extracted the small RNA population with the EasyPure miRNA kit (Transgenbiotech, China). The obtained miRNAs were measured with Agilent Bioanalyzer with the Bioanalyzer High Sensitivity RNA Analysis kit, (Agilent, USA). After checking that all the quality parameters were good, the preparation of the library for massive sequencing of the Ion Torrent platform was continued following the protocol of the Qiaseq miRNA Library Kit (Qiagen, Germany) with subsequent tempering in the IonChef (ThermoFisher, USA) with the Ion 550™ Kit-Chef kit (ThermoFisher, USA). Once the chip was loaded, it was read in the Ion S5 XL sequencer with the specific reagents included in the kit.
After sequencing the library, the miRNA sequences of each sample were obtained and used to search the following databases: miRBase v22genome-build-id: GRCh38, genome-build-accession: NCBI_Assembly: GCA_000001405. 15), Bsgnenome.Hsapiens.UCSC.hg38. masked,https://bioconductor.org/packages/release/data/annotation/html/BSgenome.Hsapiens.UCSC.hg38.masked.html (reference genome) in order to identify which miRNAs were present in each sample and how many were copies. Subsequently, a bioinformatic analysis was performed to compare the miRNAs present in each group to identify those miRNAs that provided information on the prediction of the appearance of colorectal lesions.
Once the miRNAs of interest for their overexpression or underexpression were identified, Taqman probes were designed for each of them compatible for ThermoFisher cDNA Taqman Advanced miRNA Chef chemistry (ThermoFisher, USA) which was subsequently used to test whether the same information could be obtained by real-time quantitative PCR (qPCR), since this technique is faster and cheaper. Therefore, from the preparation of urinary EVs used for the massive sequencing study, new miRNA extraction was performed with the same protocol as the one described for sequencing. With the pool of miRNAs obtained and using TaqMan probes, qPCR was performed on a QuantStudio 6 thermal cycler (ThermoFisher, USA) following the protocol described for the kit.
Results
Initially, 46 subjects were included in the study with the collection of 46 urine samples, but, due to the Covid-19 pandemic situation, only 18 samples could be analyzed, which were considered sufficient in the investigation as proof of concept. The baseline characteristics of the patients, their lifestyle and history are shown in table 1. Of the 46 subjects, 31 (69.39%) had first-degree AF CRC, 20 of them (43.47%) had no polyps or hyperplastic polyps (group 1), 11 (23.91%) had colorectal adenomas (group 2) and the rest, 15 subjects (32.60%) who were the controls, had no history of first-degree CRC and no polyps.
Table 1
Baseline characteristics, life habits, Baseline characteristics, life habits, consumption of toxins and drugs and family history of tumors of the study subjects.
All colonoscopies were complete, meeting quality standards. The preparation was adequate (Boston ≥ 6) in 43 patients (93.48%). Regarding the histology of the adenomas, only one was advanced (high-grade dysplasia).
Analysis of tetraspanins
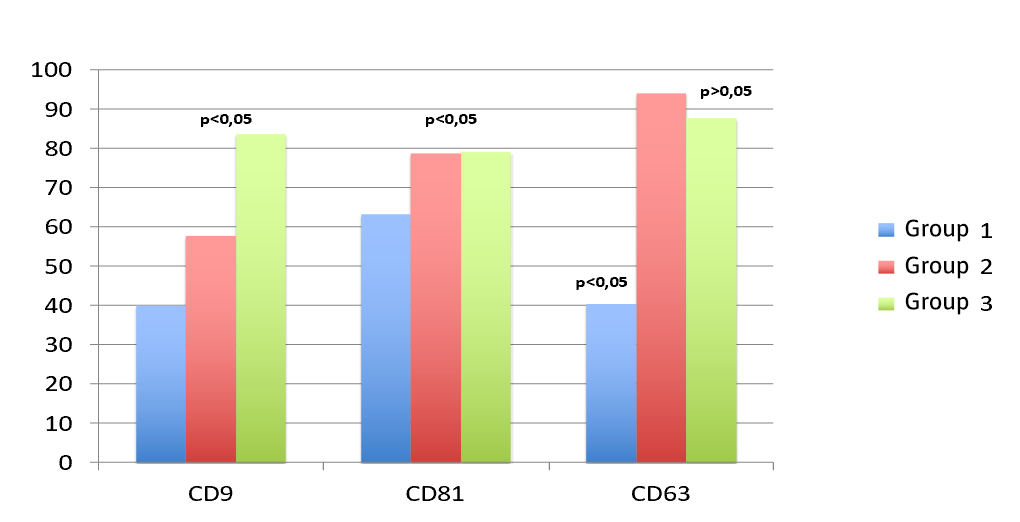
Table 2 shows the values described as percentage of positivity of CD9, CD81 and CD63 tetraspanins and figure 1 shows the comparison between the three groups.
Table 2
Percentage of positivity of tetraspanins CD9, CD81 and CD63 expressed with their mean±DE in each group.
| Group | CD9 | CD81 | CD63 |
| Group 1 | 39.87 ± 6.11 | 66.30 ± 6.87 | 40.36 ± 4.01 |
| Group 2 | 57.44 ± 5.44 | 78.75 ± 7.87 | 94.07 ± 3.99 |
| Group 3 | 83.56 ± 7.89 | 79.17 ± 6.71 | 87.66 ± 8.45 |
CD9: lower values were observed in both groups with AF CRC compared to controls, with statistically significant differences (p<0.05).
CD81: lower levels were identified in both groups with AF CCR (groups 1 and 2) compared to controls.
CD63: levels in group 1 were lower than controls, with statistically significant differences (p<0.05). In group 2, levels were higher than in controls, although without statistical differences.
Analysis of microRNAs in urinary EVs
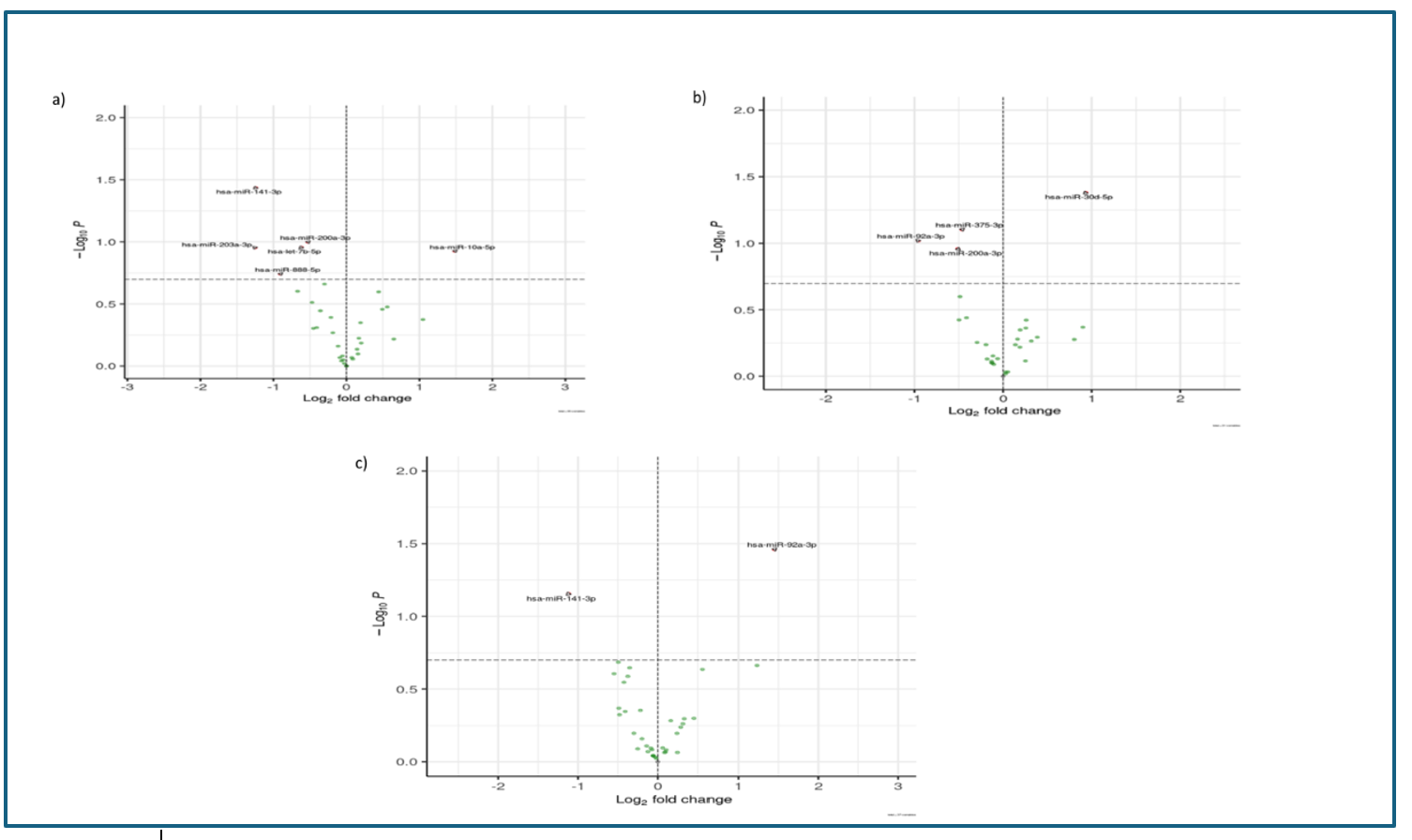
For the analysis of miRNAs present in urinary EVs, after alignment of the obtained sequences with published miRNA databases, comparison between groups was performed and represented in volcano plots (Figure 2). When comparing group 1 and group 3, 6 differentially expressed miRNAs were detected. Of note were hsa-mir-141-3p, down-expressed and with p<0.05, miR-let-7b, which is an oncogenic suppressor miRNA, also down-expressed and hsa-miR-10a-5p which is an oncogenic miRNA and found to be over-expressed.
Figure 2
Volcano plots. a) Comparison between Volcano plots. a) Comparison between group 1 and group 3. b) Comparison between group 2 and group 3. c) Comparison between group 2 and group 1.
Comparison between group 2 and group 3 allowed the detection of 4 differentially expressed miRNAs. Only hsa-miR-30d-5p showed a statistically significant elevated expression (p<0.05). The rest of miRNAs showed decreased expression in the control group of patients, with the hsa-miR-200a-3p oncomiRNA standing out.
An additional study was performed comparing both groups with AF CRC, finding hsa-miR-92-3p overexpressed in group 2. This same miRNA was also detected overexpressed when compared to controls.
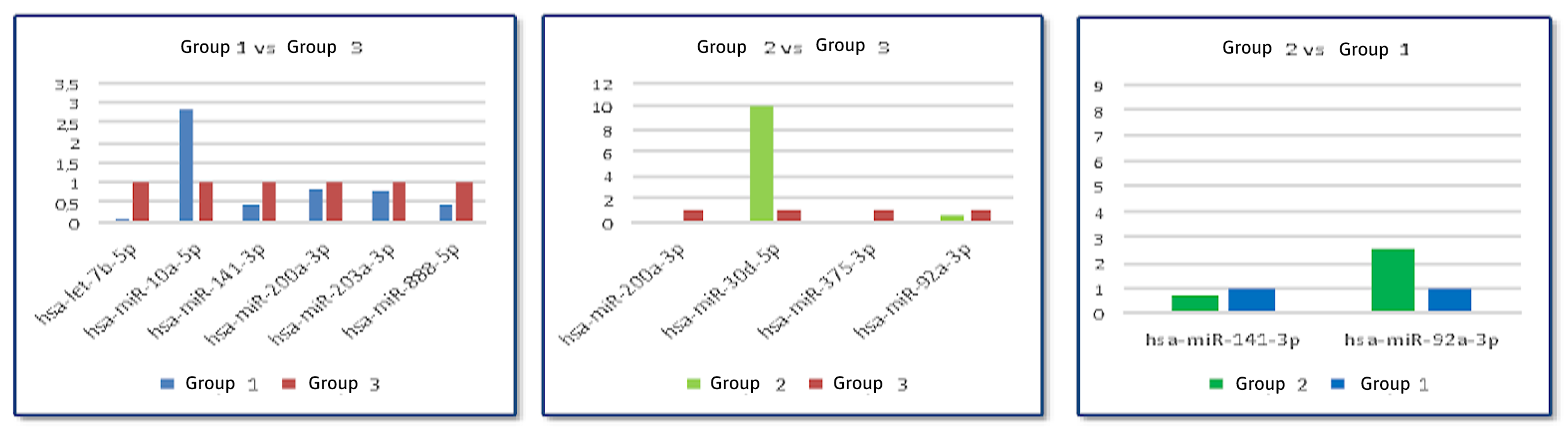
Of all the miRNAs sequenced, only 9 showed significant differences between groups (hsa-miR-92a-3p, hsa-miR-200a-3p, hsa-miR-141-3p, hsa-miR-203a-3p, hsa-miR-375-3p, hsa-miR-10a-5p, hsa-miR-30d-5p, hsa-miR-888-5p and hsa-let-7b-5). These were selected for study by qPCR using Taqman probes specific for each (Figure 3). As normalizing or "housekeeping" genes, hsa-miR-186-5p and hsa-miR-191-5p were used, since they were present in all the samples in the massive sequencing and their expression levels were maintained equally in all of them regardless of the group. The differences found in the bulk sequencing were confirmed, i.e., in the cases in which a decrease or increase of that miRNA was identified, a decrease or increase in the expression by qPCR was seen.
Discussion
In the present investigation we analyzed miRNAs carried in urine EVs, whose function is to regulate genomic transcription. A noteworthy aspect is the role of some miRNAs in the pathogenesis of adenomas and CRC. There is a growing literature on miRNAs detected in various types of biological samples. For example, in blood, miR-21, miR-92a, and miR-17-3p[13]; in tissues, an increase of 6 miRNAs from normal tissue to CRC: miR-18a, miR-18b, miR-431, miR-503, miR-1246, and miR-4417 and a decrease of 5 other miRNAs: miR-133a, miR-375, miR-378, miR-422 and miR-479[14] with 3 miRNAs: miR-21, miR-29a and miR-135b overexpressed in adenomas versus normal colonic tissue[15]; and also in feces (miR-21 and miR-106a, miR-92a and miR-106a)[16]. Regarding stool, which offers a miRNA profile very similar to that of colonic tissue, Ahmed et al.[17] proposed a panel of 12 miRNAs (miR-7, miR-17, miR-20a, miR-21, miR-92a, miR-96, miR-106a, miR-134, miR-183, miR-196a, miR-199a-3p, miR-214) with elevated expression in CRC, especially in metastatic CRC, compared to adenomas. The same authors described 8 miRNAs (miR-9, miR-29b, miR-127-5p, miR-138, miR-143, miR-146a, miR-222, miR-938) showing lower expression in CRC patients
The miRNA profile detected in our study does not allow us to ensure any individual or collective (cluster) signature defining colorectal lesion, although at least two miRNAs (miR-141-3p and miR-30d-5p) are differentially expressed in the EVs of first-degree relatives of patients with CRC compared to the control group, with differences between the two pathological groups. Furthermore, miR-92a-3p is overexpressed in patients with adenomas, whereas miR-141-3p is down-expressed (it is associated with 81 diseases in humans including, secondarily, CRC)[18]. This data is in line with several international studies that define diagnostic miRNA profiles, especially for CRC, although they do not refer to lesions with malignant potential or to relatives of CRC patients[19]-[22]. Although our work does not allow us to draw generalizable conclusions, the low number of samples is quite common in the literature, so that a study with a larger number of samples would allow us to define miRNAs with increased expression in pre- or neoplastic colorectal lesions.
The expression profile of tetraspanins (CD9, CD63 and CD81) detected by CF, which allows rapid and efficient detection, in first-degree relatives of CRC patients is different from that of the control group. In our investigation we detected a lower level of CD9 in subjects with AF CRC compared to controls. CD9 tetraspanin is involved in cell adhesion and EV uptake to colorectal tumor cells[23]. These processes are negatively regulated by tetraspanin CD9 expression on EVs. Tetraspanins CD81 and CD63 are involved in CRC progression and, moreover, it appears that the expression of both correlates with tumor invasion and metastasis. In our study, both groups with HF CRC presented lower CD81 values compared to controls, subjects with HF CRC and adenomas presented higher CD63 levels than in controls and the group with HF CRC and no adenomas showed lower levels. A study with a larger number of samples could perhaps define a proper differential profile of tetraspanins in urinary EVs for the different stages of colorectal pathology.
Conclusions
The present investigation allows us to highlight several aspects: 1) The validity of urine as a suitable diagnostic method for the study of EVs and the miRNAs carried in them. 2) The analysis of tetraspanins, which, although they are proteins used in CF as membrane markers, could complement or substitute the determination of miRNAs; that is, it could be sufficient to analyze the tetraspanin profile instead of miRNAs, which requires more time, personnel, material and economic resources as it involves genomic technology instead of CF (more versatile, cheaper and faster). And 3) This proof of concept may open a door for the possible design of a panel of Taqman probes that, by qPCR, study the expression of miRNAs in each at-risk subject in search of a non-invasive preventive diagnosis of colorectal lesions.

 Descargar número completo
Descargar número completo Download full issue
Download full issue